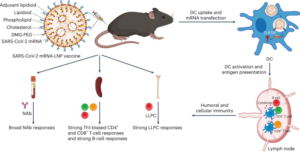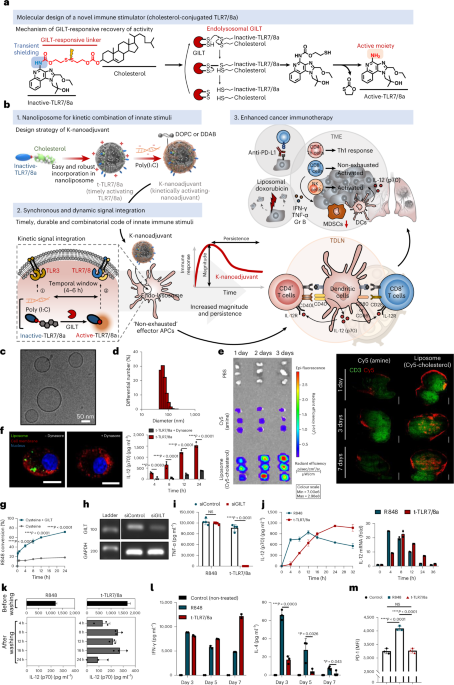
Synthesis and characterization of t-TLR7/8a and K-nanoadjuvant
Cholesterol-conjugated TLR7/8a was fabricated through the chemical scheme shown in Supplementary Figs. 1–3. The structures of the synthesized compounds were characterized by 1H NMR (Bruker Avance III, 700 MHz). For the preparation of liposomes (timely activating TLR7/8a, t-TLR7/8a), dimethyl dioctadecyl ammonium bromide (Sigma-Aldrich), 1,2-dioleoyl-sn-glycero-3-phosphocholine (Avanti Polar Lipid) and cholesterol-conjugated TLR7/8a (molar ratio, 2:5.3:1) were dissolved in chloroform/methanol (volume ratio, 9:1). The organic solvent was completely removed with a rotary evaporator at r.t. for 30 min. The thin film was hydrated with PBS, and the resulting solution was sonicated for 2 min via tip sonication in ice-bath conditions. Then, the final liposomes were extruded through filter membranes with 0.4 μm and 0.2 μm pore sizes using a Mini Extruder (Avanti Polar Lipid). As controls, blank liposomes and liposomes containing R848 (MedChemExpress) or an admixture of R848 and cholesterol (Sigma-Aldrich) were fabricated by the same method. The loaded amounts of t-TLR7/8a and free R848 were quantified by ultraviolet–visible light spectrometry (UV-1800). The hydrodynamic size and zeta potential of liposome formulations were measured using dynamic light scattering (DLS, ELS-Z electrophoretic light scattering photometer). Morphology was analysed by cryogenic transmission electron microscopy (FEI Tecnai F20 G2, Advanced Analysis Center, Korea Institute of Science and Technology).
To obtain and confirm the optimal conditions for K-nanoadjuvant, poly(I:C) was added so that the mass ratio of t-TLR7/8a to poly(I:C) (Sigma-Aldrich) was approximately 12:1. Then, poly(I:C), t-TLR7/8a and K-nanoadjuvant were run on a 1% agarose gel by electrophoresis in 1× Tris acetate–EDTA buffer (TAE, LPS solution) at 100 V for 40 min. Gel separation was visualized using a BioDoc-It imaging system (UVP).
Animals, cell lines and antibodies
The animal study was reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) of Sungkyunkwan University School of Medicine (SKKUIACUC2020-12-13-1), which is accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International and abides by the Institute of Laboratory Animal Resources guidelines. C57BL/6 and BALB/C mice (6- to 8-week-old female) were purchased from Orient Bio and DBL (Korea). IFNAR1−/− mice were received from Sang-Jun Ha (Yonsei University, Korea). OT-I mice were received from Yong-Soo Bae (Sungkyunkwan University, Korea). All animals were housed in individually ventilated cage under conditions of 30–70% humidity, 21–26 °C temperature and a 12 h light–dark cycle. RAW 264.7 cells (macrophage cell line, ATCC) and murine B16OVA tumour cells (melanoma, ATCC) were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Thermo Fisher). Murine TC-1 tumour cells (cervical cancer, ATCC) and murine 4T1 tumour cells (breast cancer, ATCC) were cultured in RPMI 1640 medium (Thermo Fisher). RAW-Blue cells (InvivoGen) were cultured in DMEM containing Zeocin (200 μg ml−1, InvivoGen). The cell lines examined in this study were used after confirming they were free of mycoplasma contamination and were not listed in the misidentified cell lines. All media were supplemented with 10% heat-inactivated fetal bovine serum (Thermo Fisher), penicillin (50 IU ml−1) and streptomycin (50 μg ml−1, Thermo Fisher). Detailed information on the antibodies such as fluorescent antibody type, manufacturer, clone and catalogue number used in this study are provided in Supplementary Table 3.
In vitro BMDC culture and cellular uptake assay
BMDCs were generated from the bone marrow of C57BL/6 mice (6- to 8-week-old female). The femurs and tibias were isolated, and the bone marrow was flushed with RPMI 1640 medium (without HEPES, Thermo Fisher Scientific) using a 26-gauge syringe. The red blood cells (RBCs) were removed with RBC lysis buffer (BioLegend). After washing, the cells were resuspended in RPMI medium (24 ml) containing mGM-CSF (20 ng ml−1, CreaGene) and seeded (2.5 × 106 per well) in a 6-well culture plate. On day 2, the medium containing mGM-CSF (20 ng ml−1) was changed after vigorous washing with PBS to remove the non-adherent cells. On day 4, fresh medium containing mGM-CSF (20 ng ml−1) was added. Differentiated immature BMDCs were used on day 6.
For cellular uptake of liposomes, liposomes were prepared with FITC–cholesterol (TopFluor Cholesterol, Avanti Polar Lipid). Immature BMDCs (4 × 104 cells) were seeded in an ibidi μ-slide 8-well microscopy chamber. The cells were preincubated with Dynasore (40 μM, Sigma-Aldrich) for 1 h and incubated with liposome(FITC–cholesterol) at 37 °C for 4 h. After washing with PBS, the cell membrane was stained with wheat germ agglutinin Texas red (Thermo Fisher), and the nuclei were stained with Hoechst 33342 (Invitrogen). Cell imaging was performed using a DeltaVision PD (GE Life Sciences) equipped with a 100× objective and the following filter sets (excitation (nm) /emission (nm)): Cy5 (645/679), FITC (490/525), TRITC (555/605) and DAPI (360/465).
GILT-dependent t-TLR7/8a analysis
GILT-dependent t-TLR7/8a cleavage
t-TLR7/8a (1.27 mM, 50 μl) was prepared in a 5 ml tube. Cysteine (1 μM or 200 nM, 50 μl, Sigma-Aldrich) was dissolved in PBS with or without GILT (2.5 μg, recombinant human IFI30, RayBiotech) and added to the tube. All samples were incubated in a 37 °C shaking incubator. Samples were snap-frozen with liquid nitrogen every hour and stored at −20 °C. After 12 h, all samples were lyophilized at −80 °C in a vacuum freeze dryer (FDU-2100, EYELA) under 10 Pa for 24 h. The lyophilized samples were analysed by liquid chromatography–mass spectrometry (Agilent 1100) to quantify the amount of R848.
GILT gene knockdown analysis
RAW 264.7 cells (2 × 105 cells per well) were seeded in a 6-well culture plate, and after 24 h, the cells were refed with fresh DMEM. Lipofectamine RNAiMAX (Thermo Fisher, 12 μl), IFI30-specific siRNA (Genolotion) and opti-MEM (Thermo Fisher) were mixed in a total volume of 300 μl, and the solution was incubated at r.t. for 5 min. The cells were treated with the solution (40 pmol per well) and incubated for 18 h. After GILT gene knockdown induction, the cells were refed with fresh medium and treated with R848 (1 μg ml−1, 3.18 μM) and t-TLR7/8a (2.9 μg ml−1, 3.18 μM). The cell supernatants were collected 24 h later, and TNF-α secretion was measured by enzyme-linked immunosorbent assay (ELISA).
In vitro Th1/Th2 polarization of CD4+ T cells
BMDCs were pretreated with R848 (1 μg ml−1, 3.18 μM) or t-TLR7/8a (2.9 μg ml−1, 3.18 μM) in the presence of OVA (10 μg ml−1) for 12 h. Spleens were harvested from C57BL/6 mice, and CD4+ T cells were isolated using a naive CD4+ T-cell isolation kit, mouse (Miltenyi Biotec) according to the manufacturer’s protocol. Preincubated DCs were co-cultured with CD4+ T cells (DC:T-cell ratio = 1:10) in a 96-well flat-bottom plate. After 3, 5 or 7 days, the cell culture supernatants were collected, and IL-4 and IFN-γ secretion was measured by ELISA.
Histological analysis
Naive C57BL/6 mice (6-week-old female) were subcutaneously injected with R848 (50 μg, 159 μmol), poly(I:C) (12.5 μg), t-TLR7/8a (144.2 μg, 159 μmol), R848 + poly(I:C) or K-nanoadjuvant four times every 3 days. Three days after the last injection, the liver, lung, spleen and kidney were harvested. The samples were sectioned and stained with H&E. The stained sections were visualized by inverted microscopy (Eclipse Ts2, A-FRONTIER) with a 40× objective.
In vivo flow cytometry analysis
B16OVA or TC-1 tumour cells (5 × 105 cells per mouse) were subcutaneously inoculated into the right flanks of C57BL/6 mice (6-week-old female). After randomly distributing tumour-bearing mice to groups 5–7 days later, R848 (25 μg, 79.5 μmol), poly(I:C) (6.25 μg), t-TLR7/8a (72.1 μg, 79.5 μmol), R848 + poly(I:C), or K-nanoadjuvant with SIINFEKL (15 μg, OVA257-264 peptide antigen, MIMOTOPES) or the E7-long peptide (AGQAEPDRAHYNIVTFCCKCDS) (10 μg, Anygen) was administered. All immunizations were repeated every 3 days for a total of three times. The non-treated group was used as a control.
Preparation of single-cell suspensions
Tumours and TDLNs were mechanically disrupted and resuspended in medium containing collagenase D (1 mg ml−1, Sigma-Aldrich). The solutions were incubated in a shaking incubator for 40 min at 37 °C. Then, the cells were washed twice with PBS after filtration through 70 μm cell strainers. To isolate splenocytes, the spleens were mechanically homogenized and resuspended in RBC lysis buffer (BioLegend) to remove RBCs. The solutions were filtered through a 70 μm cell strainer, and medium was added. Splenocytes were obtained after the suspensions were centrifuged at 488g for 3 min.
DC activation and tumor-infiltrating leukocytes (TIL) population
The single cells from TDLNs and tumours were stained with antibodies specific for activated DCs (anti-mouse CD11c, CD80 and CD86). The single cells from tumours were stained with antibodies specific for MDSCs (anti-mouse CD11b and Gr-1) and T cells and NK cells (anti-mouse CD45, CD3, CD8, NK1.1 and CD69). Detailed information on the antibodies used is provided in Supplementary Table 3.
Peptide restimulation of CD8+ T cells and stimulation of NK cells
For antigen-specific CD8+ T-cell analysis, single cells (5 × 105 per well) were seeded in a round-bottom 96-well plate. Then, the cells were restimulated with the OVA SIINFEKL peptide (10 μg ml−1) or Long-E7 peptide (10 μg ml−1), IL-2 (30 ng ml−1, PeproTech), and GolgiPlug or GolgiStop (protein transport inhibitor, 0.6 μg ml−1, BD Bioscience) for 12 h. For NK-cell stimulation analysis, single cells (5 × 105 per well) were seeded in a round-bottom 96-well plate. The cells were stimulated with a cell activation cocktail (with brefeldin A) (BioLegend, 500X), which is a mixture of optimized concentrations of PMA, ionomycin and a protein transport inhibitor (brefeldin A) for 4 h. After stimulation, the cells were collected and washed twice. Cells were stained with surface marker antibodies for 30 min at 4 °C. Single cells from TDLNs and tumours were stained with antibodies specific for exhausted CD8+ T cells (anti-mouse CD3, CD8, PD-1, LAG-3 and TIM-3) or activated CD8+ T cells (anti-mouse CD3, CD8, CD69, 4-1BB and CD25). Detailed information on the antibodies used is provided in Supplementary Table 3.
For intracellular staining, the cells were washed and then resuspended in fixation/permeabilization solution for 20 min at 4 °C. Fixed cells were washed twice with BD Perm/Wash buffer (BD Bioscience) and stained with antibodies for 30 min at 4 °C. Single cells from TDLNs, tumours and splenocytes were stained with antibodies specific for cytokine-producing CD8+ T cells (anti-mouse CD3, CD8, IFN-γ, TNF-α and Granzyme B) or cytokine-producing NK cells (anti-mouse CD3, NK1.1, IFN-γ, TNF-α and Granzyme B). After staining, the cells were washed twice with BD Perm/Wash buffer and resuspended in staining buffer. Flow cytometry data were analysed using a BD FACSCanto II (at the BIORP of the Korea Basic Science Institute) and quantified using FlowJo v.10. Detailed information on the antibodies and gating strategies used are provided in Supplementary Table 3 and Supplementary Figs. 15, 23, 24, 38 and 39.
In vivo cytokine secretion in tumours and TDLNs
For cytokine secretion analysis in tissues, TDLNs were mechanically disrupted and resuspended in a medium containing collagenase D (1 mg ml−1, Sigma-Aldrich). Single-cell suspensions obtained from the TDLNs were seeded in a 96-well round-bottom culture plate and incubated at 37 °C for 24 h. The cells were harvested, and the supernatants were collected after centrifugation. The tumours were suspended in CellLytic MT cell lysis reagent (100 mg tissue per ml, Sigma-Aldrich) and mechanically disrupted. The solutions were centrifuged at 10,000g for 10 min at 4 °C, and the supernatants were collected. All collected supernatants were analysed with IL-12(p70) or IFN-γ ELISA kits.
In vivo kinetics of IL-12(p70) cytokine secretion and immune cell numbers in LNs
To analyse the kinetics of IL-12(p70) cytokine secretion in LNs, naive C57BL/6 mice received a single injection of poly(I:C) (6.25 μg) + R848 (25 μg, 79.5 μmol) or t-TLR7/8a (72.1 μg, 79.5 μmol) with SIINFEKL (15 μg). The draining inguinal LNs were harvested at serial time points (6, 12, 24, 48 and 72 h). Then, we proceed in the same way as described above. All collected supernatants were analysed with IL-12(p70) or IFN-γ ELISA kits. To analyse the kinetics of immune cell numbers in the LNs, naive C57BL/6 mice received a single injection equal to the above dose without antigen. The draining inguinal LNs were harvested at serial time points (0, 1, 2, 4, 7 and 14 days). Then, we proceeded in the same way as described above to obtain single cells from the lymph node above.
In vivo antitumour study
TC-1 tumour cells (5 × 105 cells per mouse) were subcutaneously inoculated into the right flanks of C57BL/6 mice (6-week-old female). Poly(I:C) (6.25 μg) + R848 (25 μg, 79.5 μmol) or t-TLR7/8a (72.1 μg, 79.5 μmol) with E7-long peptide (10 μg) was injected according to the indicated schedule after random distribution of tumour-bearing mice to the groups. The PBS-treated group was used as a control. Mouse tumour growth and survival were monitored at various time points. The tumour volume was calculated using the following formula: (long-axis diameter) × (short-axis diameter)2/2. The mice were killed when the tumour volume reached the maximum tumour size (1,000 mm3) approved by the IACUC, Sungkyunkwan University School of Medicine.
Combination with immune checkpoint blockade
After 4 days of tumour inoculation, the mice received K-nanoadjuvant (t-TLR7/8a, 72.1 μg; poly(I:C), 6.25 μg) with the E7-long peptide (10 μg), and immunizations were performed every 3 days for a total of six times. Anti-PD-L1 antibodies (clone: 10 F. G2, BioXCell, 100 μg) were administered intraperitoneally every 2 days for a total of eight times.
Combination with chemotherapy
After 4 days of tumour inoculation, mice were injected with K-nanoadjuvant (t-TLR7/8a, 72.1 μg; poly(I:C), 6.25 μg) with the E7-long peptide (10 μg), and immunizations were repeated five times every 3 days. Liposome(doxorubicin) was fabricated by the method described in the Supplementary Methods. Mice were injected with liposome(doxorubicin) (80 μg of doxorubicin) two times at 6-day intervals.
In vivo IL-12 depletion study
B16OVA tumour cells (5 × 105 cells per mouse) were subcutaneously inoculated into the right flanks of mice. The mice received a subcutaneous injection of K-nanoadjuvant (t-TLR7/8a, 72.1 μg; poly(I:C), 6.25 μg) with SIINFEKL (15 μg) three times every 3 days. Beginning 3 days before the first injection of K-nanoadjuvant, anti-mouse IL-12p75 (clone: R2-9A5, BioXcell, 300 μg) was administered intraperitoneally for a total of five times at 3-day intervals.
In vivo distant tumour model study
TC-1 tumour cells (5 × 105 cells per mouse) were subcutaneously inoculated into the right flanks of C57BL/6 mice (6-week-old female) on day 0. At 4 days after primary tumour inoculation, secondary tumour cells (2.5 × 105 cells per mouse) were subcutaneously inoculated in the left flank. K-nanoadjuvant (t-TLR7/8a, 72.1 μg; poly(I:C), 6.25 μg) was injected four times every 3 days beginning 4 days after primary tumour inoculation. Mouse tumour growth was monitored at various time points. On day 22, the tumours were harvested and photographed.
In vivo orthotopic TC-1 model study
C57BL/6 mice (6-week-old female) were anaesthetized via inhaled anaesthesia. The right side of the thorax skin was incised. Then, the mice were inoculated with TC-1 cells (5 × 105 cells per mouse) on the right side of the lung lobe. After 3 days, the K-nanoadjuvant (t-TLR7/8a, 72.1 μg, 79.5 μmol; poly(I:C), 6.25 μg) and R848 (25 μg, 79.5 μmol) + poly(I:C) (6.25 μg) groups were subcutaneously administered, and immunizations were performed every 3 days for a total of four times. On day 14, the lungs were harvested, sectioned, and stained with H&E. The H&E-stained sections were visualized by using a microscope (Zeiss Axiovert 200 M) equipped with a 40× objective. The control group and the R848 + poly(I:C) and K-nanoadjuvant groups were killed on days 14 and 31, respectively, for lung staining.
Lung metastasis analysis
BALB/C mice (6-week-old female) were inoculated in the right flank with 4T1 tumour cells (5 × 105 cells per mouse). After 5 days, the mice were administered K-nanoadjuvant (t-TLR7/8a, 72.1 μg; poly(I:C), 6.25 μg) with tumour lysate (10 μg), and immunization was performed every 3 days for a total of four times. After approximately 1 month, lung metastasis was evaluated using India ink (3 ml in 47 ml of PBS), which was administered by intratracheal injection. The lungs were excised, washed with PBS, and immersed in a fixative solution (70% ethanol (40 ml), 4% formaldehyde (1 ml), and acetic acid (0.5 ml)). Lung metastatic nodules were counted by visual observation.
In vivo tumour rechallenge model study
TC-1 tumour cells (5 × 105 cells per mouse) were subcutaneously inoculated into the right flanks of C57BL/6 mice (6-week-old female) on day 0. After 4 days, mice were injected with K-nanoadjuvant five times every 3 days, and the mice were injected with anti-PD-L1 antibodies (100 μg) five times every 2 days beginning 7 days after tumour inoculation. On day 17, the mice were killed and TDLNs and spleen were collected. Single cells from TDLNs and splenocytes were stained with antibodies specific for memory T cells (anti-mouse CD3, CD8, CD44 and CD62L). Detailed information on the antibodies used is provided in Supplementary Table 3. Flow cytometry was then performed using a BD FACSCanto II to analyse the stained cell suspensions. Twenty-one days after tumour inoculation, naive mice and K-nanoadjuvant-treated tumour-free mice were rechallenged with TC-1 tumour cells (5 × 105 cells per mouse) in the same right flank.
Statistics and reproducibility
All results are indicated as the mean ± s.d. A two-tailed unpaired t-test was used to compare the two groups. One-way analysis of variance (ANOVA) (or two-way ANOVA) with Tukey’s multiple comparisons test (or Sidak’s multiple comparisons test) was used to analyse multiple groups of data. The log-rank (Mantel–Cox) test was used for survival data. All statistical analyses were performed using GraphPad Prism 8 and Microsoft Excel 2016. P values (NS, not significant, *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001) were used to indicate statistical significance. Experiments in Figs. 1c,e,f,h and 2j were repeated three times taken from distinct samples. Experiments in Figs. 2i and 5a,f were repeated two times taken from distinct samples.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
- SEO Powered Content & PR Distribution. Get Amplified Today.
- Platoblockchain. Web3 Metaverse Intelligence. Knowledge Amplified. Access Here.
- Source: https://www.nature.com/articles/s41565-022-01296-w
- 000
- 1
- 10
- 100
- 2016
- 7
- 70
- 9
- a
- above
- According
- accreditation
- accredited
- activating
- Activation
- added
- administered
- advanced
- After
- All
- amount
- amounts
- analyse
- analysis
- Anchor
- and
- animal
- animals
- Antibodies
- approved
- approximately
- article
- assessment
- Association
- available
- Avanti
- basic
- BD
- before
- Beginning
- blood
- BONE
- Breast cancer
- buffer
- calculated
- Cancer
- care
- Cells
- Center
- Chamber
- characterized
- chemical
- cocktail
- compare
- completely
- conditions
- Confirm
- control
- controls
- cryogenic
- Culture
- data
- day
- Days
- dc
- described
- Design
- detailed
- differentiated
- Distant
- distinct
- distributing
- distribution
- dryer
- dynamic
- Enhances
- equipped
- evaluated
- Excel
- fei
- female
- Figure
- Film
- filter
- final
- First
- fixed
- flow
- Flushed
- following
- formaldehyde
- formula
- Free
- Freeze
- fresh
- from
- G2
- ge
- generated
- Group
- Group’s
- Growth
- guidelines
- homogenized
- HTTPS
- human
- Imaging
- immersed
- in
- incubated
- incubator
- india
- indicate
- Individually
- information
- innate
- Institute
- Institutional
- International
- isolated
- isolation
- kidney
- kit
- korea
- laboratory
- Last
- Life
- Life Sciences
- light
- Line
- lines
- LINK
- linked
- Liquid
- Listed
- Liver
- LPs
- Lungs
- Manufacturer
- marker
- Mass
- material
- maximum
- Media
- medicine
- medium
- Memory
- method
- mice
- Microscope
- Microscopy
- Microsoft
- Microsoft Excel
- mixed
- mixture
- ML
- model
- modified
- monitored
- Month
- MT
- multiple
- Nature
- NK
- node
- number
- numbers
- objective
- obtained
- optimal
- optimized
- organic
- PBS
- plato
- Plato Data Intelligence
- PlatoData
- points
- polar
- portfolio
- potential
- prepared
- presence
- primary
- Protein
- protocol
- provided
- purchased
- random
- ratio
- Raw
- rbc
- reached
- received
- Red
- reduces
- remove
- Removed
- repeated
- Reporting
- research
- Resources
- resulting
- Results
- reviewed
- Run
- same
- schedule
- scheme
- School
- Science
- Science and Technology
- SCIENCES
- secondary
- sections
- serial
- Serum
- Sets
- shown
- significance
- significant
- single
- SIX
- Size
- sizes
- Skin
- So
- solution
- Solutions
- specific
- statistical
- stored
- strategies
- Study
- such
- Surface
- suspended
- Suspensions
- system
- T cells
- table
- Technology
- test
- texas
- The
- three
- Through
- time
- times
- tip
- tissues
- to
- Total
- transport
- under
- university
- use
- Vacuum
- Values
- various
- via
- vivo
- volume
- which
- without
- zephyrnet
- Zeta