The role of natural products in modern drug discovery.
An. Acad. Bras. Cienc. 2019; 91e20190105
A trade-off between plant and soil carbon storage under elevated CO2.
Nature. 2021; 591: 599-603
Towards food security: current state and future prospects of agrobiotechnology.
Trends Biotechnol. 2018; 36: 1219-1229
Synthetic biology tools to engineer microbial communities for biotechnology.
Trends Biotechnol. 2019; 37: 181-197
Systems and synthetic biology tools for advanced bioproduction hosts.
Curr. Opin. Biotechnol. 2020; 64: 101-109
Synthetic development: building mammalian multicellular structures with artificial genetic programs.
Curr. Opin. Biotechnol. 2019; 59: 130-140
Engineering advanced cancer therapies with synthetic biology.
Nat. Rev. Cancer. 2019; 19: 187-195
Principles of genetic circuit design.
Nat. Methods. 2014; 11: 508-520
Synthetic biology in plants, a boon for coming decades.
Mol. Biotechnol. 2021; 63: 1138-1154
Plant-based engineering for production of high-valued natural products.
Nat. Prod. Rep. 2022; 39: 1492-1509
Contemporary tools for regulating gene expression in bacteria.
Trends Biotechnol. 2020; 38: 316-333
Synthetic switches and regulatory circuits in plants.
Plant Physiol. 2019; 179: 862-884
Transcriptional and post-transcriptional controls for tuning gene expression in plants.
Curr. Opin. Plant Biol. 2023; 71102315
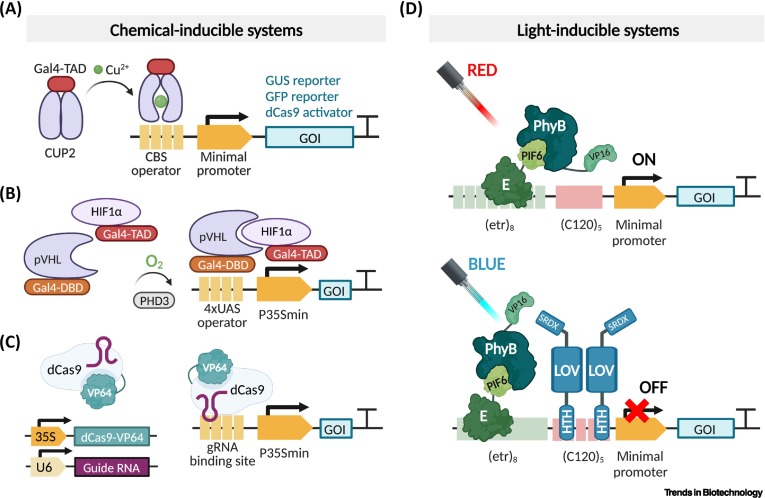
Copper-controllable gene expression system for whole plants.
Proc. Natl. Acad. Sci. U. S. A. 1993; 90: 4567-4571
Development of a tightly regulated and highly responsive copper-inducible gene expression system and its application to control of flowering time.
Plant Cell Rep. 2014; 33: 47-59
A copper switch for inducing CRISPR/Cas9-based transcriptional activation tightly regulates gene expression in Nicotiana benthamiana.
BMC Biotechnol. 2022; 22: 1-13
A synthetic oxygen sensor for plants based on animal hypoxia signaling.
Plant Physiol. 2019; 179: 986-1000
Applications of CRISPR–Cas in agriculture and plant biotechnology.
Nat. Rev. Mol. Cell Biol. 2020; 21: 661-677
Orthogonal control of gene expression in plants using synthetic promoters and CRISPR-based transcription factors.
Plant Methods. 2022; 18: 42
The impact of inducible promoters in transgenic plant production and crop improvement.
Plant Gene. 2021; 27: 100300
Synthetic biology approaches in regulation of targeted gene expression.
Curr. Opin. Plant Biol. 2021; 63102036
The trans-regulatory landscape of gene networks in plants.
bioRxiv. 2022; (Published online October 24, 2022. https://doi.org/10.1101/2022.10.23.513368)
Advances in engineering and optimization of transcription factor-based biosensors for plug-and-play small molecule detection.
Curr. Opin. Biotechnol. 2022; 76102753
Plant optogenetics: applications and perspectives.
Curr. Opin. Plant Biol. 2022; 68102256
Optogenetic control of gene expression in plants in the presence of ambient white light.
Nat. Methods. 2020; 17: 717-725
Development of biosensors and their application in metabolic engineering.
Curr. Opin. Chem. Biol. 2015; 28: 1-8
Aptamer-based and aptazyme-based riboswitches in mammalian cells.
Curr. Opin. Chem. Biol. 2019; 52: 72-78
Inducible gene expression from the plastid genome by a synthetic riboswitch.
Proc. Natl. Acad. Sci. U. S. A. 2010; 107: 6204-6209
Boosting riboswitch efficiency by RNA amplification.
Nucleic Acids Res. 2015; 43e66
Riboswitch-mediated inducible expression of an astaxanthin biosynthetic operon in plastids.
Plant Physiol. 2022; 188: 637-652
A theophylline-responsive riboswitch regulates expression of nuclear-encoded genes.
Plant Physiol. 2020; 182: 123-135
Highly specific gene silencing by artificial microRNAs in Arabidopsis.
Plant Cell. 2006; 18: 1121-1133
Recent advances in plant gene silencing methods.
Plant Gene Silencing. 2022; 2408: 1-22
Micro-RNA based gene regulation: a potential way for crop improvements.
Plant Gene. 2021; 27100312
Cis-trans recognition and subunit-specific degradation of short-lived proteins.
Nature. 1990; 346: 287-291
Phenotypes on demand via switchable target protein degradation in multicellular organisms.
Nat. Commun. 2016; 7: 1-15
Modulating protein stability to switch toxic protein function on and off in living cells.
Plant Physiol. 2019; 179: 929-942
Selectable markers and reporter genes: a well furnished toolbox for plant science and genetic engineering.
CRC Crit. Rev. Plant Sci. 2012; 31: 401-453
A reporter for noninvasively monitoring gene expression and plant transformation.
Hortic. Res. 2020; 7: 152
Building customizable auto-luminescent luciferase-based reporters in plants.
Elife. 2020; 9e52786
Construction of a genetic toggle switch in Escherichia coli.
Nature. 2000; 403: 339-342
A synthetic oscillatory network of transcriptional regulators.
Nature. 2000; 403: 335-338
Robust multicellular computing using genetically encoded NOR gates and chemical ‘wires’.
Nature. 2011; 469: 212-215
Synthetic circuits integrating logic and memory in living cells.
Nat. Biotechnol. 2013; 31: 448-452
Using synthetic biology to engineer spatial patterns.
Adv. Biosyst. 2019; 3e1800280
A memory switch for plant synthetic biology based on the phage φc31 integration system.
Nucleic Acids Res. 2020; 48: 3379-3394
An integrase toolbox to record gene-expression during plant development.
bioRxiv. 2022; (Published online September 17, 2022. https://doi.org/10.1101/2022.09.16.508262)
Synthetic memory circuits for stable cell reprogramming in plants.
Nat. Biotechnol. 2022; 40: 1862-1872
CRISPRi-based circuits for genetic computation in plants.
bioRxiv. 2022; (Published online July 1, 2022. https://doi.org/10.1101/2022.07.01.498372)
Synthetic genetic circuits as a means of reprogramming plant roots.
Science. 2022; 377: 747-751
The second decade of synthetic biology: 2010–2020.
Nat. Commun. 2020; 11: 1-4
Developing a Genetic Toggle Switch in Arabidopsis.
American Physical Society, 2022
A tunable synthetic mammalian oscillator.
Nature. 2009; 457: 309-312
Intracellular gate opening in Shaker K+ channels defined by high-affinity metal bridges.
Nature. 2004; 428: 864-868
Characterization and mitigation of gene expression burden in mammalian cells.
Nat. Commun. 2020; 11: 1-11
Spatiotemporal control of gene expression with pulse-generating networks.
Proc. Natl. Acad. Sci. U. S. A. 2004; 101: 6355-6360
Fold-change detection and scalar symmetry of sensory input fields.
Proc. Natl. Acad. Sci. U. S. A. 2010; 107: 15995-16000
Genetic circuit design automation with Cello 2.0.
Nat. Protoc. 2022; 17: 1097-1113
Genetic circuit design automation for yeast.
Nat. Microbiol. 2020; 5: 1349-1360
Precise, automated control of conditions for high-throughput growth of yeast and bacteria with eVOLVER.
Nat. Biotechnol. 2018; 36: 614-623
Engineering microbial consortia for controllable outputs.
ISME J. 2016; 10: 2077-2084
Automated design of synthetic microbial communities.
Nat. Commun. 2021; 12: 672
Quantitative and predictive genetic parts for plant synthetic biology.
Front. Plant Sci. 2020; 11512526
A general strategy to construct small molecule biosensors in eukaryotes.
Elife. 2015; 4e10606
Rational design of minimal synthetic promoters for plants.
Nucleic Acids Res. 2021; 48: 11845-11856
Design of orthogonal regulatory systems for modulating gene expression in plants.
Nat. Chem. Biol. 2020; 16: 857-865
Toolboxes for plant systems biology research.
Curr. Opin. Biotechnol. 2022; 75102692
DNA assembly for plant biology: techniques and tools.
Curr. Opin. Plant Biol. 2014; 19: 14-19
Phytobricks: manual and automated assembly of constructs for engineering plants.
DNA Clon. Assembly. 2020; 2205: 179-199
An extensible vector toolkit and parts library for advanced engineering of plant genomes.
Plant Genome. 2022; (Published online March 9, 2023. https://doi.org/10.1002/tpg2.20312)e20312
Standards for plant synthetic biology: a common syntax for exchange of DNA parts.
New Phytol. 2015; 208: 13-19
Quantitative characterization of genetic parts and circuits for plant synthetic biology.
Nat. Methods. 2015; 13: 94-100
Building the plant SynBio toolbox through combinatorial analysis of DNA regulatory elements.
ACS Synth. Biol. 2022; 11: 2741-2755
Construction of DNA Tools for hyperexpression in Marchantia chloroplasts.
ACS Synth. Biol. 2021; 10: 1651-1666
Hairy root culture technology: applications, constraints and prospect.
Appl. Microbiol. Biotechnol. 2021; 105: 35-53
Plant viruses in plant molecular pharming: toward the use of enveloped viruses.
Front. Plant Sci. 2019; 10: 803
Merging automation and fundamental discovery into the design–build–test–learn cycle of nontraditional microbes.
Trends Biotechnol. 2022; 40: 1148-1159
A robotic platform for high-throughput protoplast isolation and transformation.
J. Vis. Exp. 2016; 115e54300
Biofoundry-assisted expression and characterization of plant proteins.
Synth. Biol. 2021; 6: ysab029
The emerging role of nanotechnology in plant genetic engineering.
Nat. Rev. Bioeng. 2023; (Published online February 22, 2023. https://doi.org/10.1038/s44222-023-00037-5): 1-15
Benchmarking intrinsic promoters and terminators for plant synthetic biology research.
BioDesign Res. 2022; 2022: 9834989
Emerging opportunities for synthetic biology in agriculture.
Genes. 2018; 9: 341
The impact of synthetic biology for future agriculture and nutrition.
Curr. Opin. Biotechnol. 2020; 61: 102-109
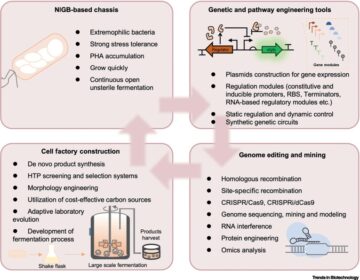
Recent trends in metabolic engineering of microbial chemical factories.
Curr. Opin. Biotechnol. 2019; 60: 188-197
Microbial chassis development for natural product biosynthesis.
Trends Biotechnol. 2020; 38: 779-796
Edible vaccines: promises and challenges.
Mol. Biotechnol. 2020; 62: 79-90
Production of antibodies in transgenic plants.
Nature. 1989; 342: 76-78
Plant molecular farming for the production of valuable proteins – critical evaluation of achievements and future challenges.
J. Plant Physiol. 2021; 258153359
Recent advances in molecular farming using monocot plants.
Biotechnol. Adv. 2022; 58107913
Plant synthetic biology innovations for biofuels and bioproducts.
Trends Biotechnol. 2022; 40: 1454-1468
Optimization of heterologous glucoraphanin production in planta.
ACS Synth. Biol. 2022; 1: 1865-1873
Reconstitution of monoterpene indole alkaloid biosynthesis in genome engineered Nicotiana benthamiana.
Commun. Biol. 2022; 5: 949
Production of volatile moth sex pheromones in transgenic Nicotiana benthamiana plants.
BioDesign Res. 2021; 2021: 9891082
Tunable control of insect pheromone biosynthesis in Nicotiana benthamiana.
bioRxiv. 2022; (June 15, 2022)
Applications of multi-omics technologies for crop improvement.
Front. Plant Sci. 2021; 12: 1846
Understanding omics driven plant improvement and de novo crop domestication: some examples.
Front. Genet. 2021; 12: 415
Proteases of Nicotiana benthamiana: an emerging battle for molecular farming.
Curr. Opin. Biotechnol. 2020; 61: 60-65
Deciphering the molecular basis of tissue-specific gene expression in plants: can synthetic biology help?.
Curr. Opin. Plant Biol. 2022; 68102241
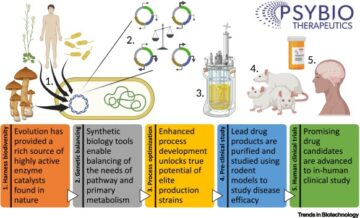
Utilizing plant synthetic biology to improve human health and wellness.
Front. Plant Sci. 2021; 12691462
Broadening the impact of plant science through innovative, integrative, and inclusive outreach.
Plant Direct. 2021; 5e00316
A steroid-inducible gene expression system for plant cells.
Proc. Natl. Acad. Sci. U. S. A. 1991; 88: 10421-10425
A glucocorticoid-mediated transcriptional induction system in transgenic plants.
Plant J. 1997; 11: 605-612
A transcription activation system for regulated gene expression in transgenic plants.
Proc. Natl. Acad. Sci. U. S. A. 1998; 95: 376-381
New pOp/LhG4 vectors for stringent glucocorticoid-dependent transgene expression in Arabidopsis.
Plant J. 2005; 41: 899-918
Stringent repression and homogeneous de-repression by tetracycline of a modified CaMV 35S promoter in intact transgenic tobacco plants.
Plant J. 1992; 2: 397-404
A chimeric transactivator allows tetracycline-responsive gene expression in whole plants.
Plant J. 1994; 5: 559-569
Ecdysone agonist inducible transcription in transgenic tobacco plants.
Plant J. 1999; 19: 97-106
An ethanol inducible gene switch for plants used to manipulate carbon metabolism.
Nat. Biotechnol. 1998; 16: 177-180
An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants.
Plant J. 2000; 24: 265-273
Novel pristinamycin-responsive expression systems for plant cells.
Biotechnol. Bioeng. 2001; 74: 154-163
Chemical-inducible, ecdysone receptor-based gene expression system for plants.
Transgenic Res. 2003; 12: 101-109
Use of bacterial quorum-sensing components to regulate gene expression in plants.
Plant Physiol. 2006; 140: 1205-1212
Programmable ligand detection system in plants through a synthetic signal transduction pathway.
PLoS One. 2011; 6e16292
A red light-controlled synthetic gene expression switch for plant systems.
Mol. BioSyst. 2014; 10: 1679-1688
A general strategy to construct small molecule biosensors in eukaryotes.
Elife. 2015; 4e10606
Computational design of environmental sensors for the potent opioid fentanyl.
Elife. 2017; 6e28909
A green-light-responsive system for the control of transgene expression in mammalian and plant cells.
ACS Synth. Biol. 2018; 7: 1349-1358
First plant-made biologic approved.
Nat. Biotechnol. 2012; 30: 472-473
Plants as factories for human pharmaceuticals: applications and challenges.
Int. J. Mol. Sci. 2015; 16: 28549-28565
Lettuce-produced hepatitis C virus E1E2 heterodimer triggers immune responses in mice and antibody production after oral vaccination.
Plant Biotechnol. J. 2017; 15: 1611-1621
Safety and immunogenicity of a plant-produced Pfs25 virus-like particle as a transmission blocking vaccine against malaria: a phase 1 dose-escalation study in healthy adults.
Vaccine. 2018; 36: 5865-5871
A plant-produced vaccine protects mice against lethal West Nile virus infection without enhancing Zika or dengue virus infectivity.
Vaccine. 2018; 36: 1846-1852
Cold chain and virus-free oral polio booster vaccine made in lettuce chloroplasts confers protection against all three poliovirus serotypes.
Plant Biotechnol. J. 2019; 17: 1357-1368
A novel anti-HIV-1 bispecific bNAb-lectin fusion protein engineered in a plant-based transient expression system.
Plant Biotechnol. J. 2019; 17: 1646-1656
Regulatory approval and a first-in-human phase I clinical trial of a monoclonal antibody produced in transgenic tobacco plants.
Plant Biotechnol. J. 2015; 13: 1106-1120
Plant-made dengue virus-like particles produced by co-expression of structural and non-structural proteins induce a humoral immune response in mice.
Plant Biotechnol. J. 2021; 19: 745-756
Immunization with plant-derived multimeric H5 hemagglutinins protect chicken against highly pathogenic avian influenza virus H5N1.
Vaccines. 2020; 8: 593
Developments in the treatment of Fabry disease.
J. Inherit. Metab. Dis. 2020; 43: 908-921
Long-lasting stable expression of human LL-37 antimicrobial peptide in transgenic barley plants.
Antibiotics. 2021; 10: 898
Efficacy and safety of a recombinant plant-based adjuvanted covid-19 vaccine.
N. Engl. J. Med. 2022; 386: 2084-2096
- SEO Powered Content & PR Distribution. Get Amplified Today.
- Platoblockchain. Web3 Metaverse Intelligence. Knowledge Amplified. Access Here.
- Source: https://www.cell.com/trends/biotechnology/fulltext/S0167-7799(23)00088-4?rss=yes
- ][p
- 1
- 10
- 2022
- 2023
- 9
- a
- achievements
- Activation
- adults
- advanced
- advances
- After
- against
- agriculture
- AL
- All
- allows
- Ambient
- Amplification
- analysis
- and
- animal
- Antibodies
- Application
- applications
- approaches
- approval
- approved
- artificial
- AS
- Assembly
- authors
- Automated
- Automation
- Bacteria
- based
- basis
- Battle
- between
- biology
- biotechnology
- blocking
- bridges
- Building
- burden
- by
- CAN
- Cancer
- carbon
- Cells
- chain
- challenges
- channels
- chassis
- chemical
- Clinical
- co2
- coming
- Common
- Communities
- components
- computation
- computing
- conditions
- consortia
- constraints
- construct
- control
- controls
- Copper
- COVID-19
- critical
- crop
- Culture
- Current
- Current state
- customizable
- cycle
- decade
- decades
- defined
- Demand
- Design
- Detection
- Development
- direct
- discovery
- Disease
- dna
- driven
- drug
- drug discovery
- during
- e
- efficiency
- elements
- elevated
- emerging
- engineer
- Engineering
- enhancing
- environmental
- estrogen
- evaluation
- examples
- exchange
- factories
- factors
- farming
- February
- Fields
- food
- For
- from
- function
- fundamental
- fusion
- future
- Gates
- General
- genetic engineering
- genome
- Growth
- Health
- healthy
- help
- highly
- hosts
- HTTPS
- human
- i
- Impact
- improve
- improvement
- improvements
- in
- Inclusive
- infection
- Influenza
- innovations
- innovative
- input
- Integrating
- integration
- intrinsic
- isolation
- ITS
- jpg
- July
- landscape
- Library
- light
- List
- living
- made
- malaria
- manual
- March
- means
- Memory
- metal
- methods
- mice
- minimal
- mitigation
- Modern
- modified
- MOL
- molecular
- molecule
- monitoring
- nanotechnology
- Natural
- network
- networks
- novel
- Novo
- nutrition
- october
- of
- on
- ONE
- online
- opening
- opportunities
- optimization
- outreach
- Oxygen
- particle
- parts
- patterns
- perspectives
- pharmaceuticals
- phase
- physical
- plants
- platform
- plato
- Plato Data Intelligence
- PlatoData
- potential
- presence
- Produced
- Product
- Production
- Products
- Programs
- promises
- promoters
- prospect
- prospects
- protect
- protection
- Protein
- Proteins
- recognition
- record
- Red
- Regulate
- regulated
- Regulation
- Regulators
- regulatory
- reporter
- research
- response
- responsive
- RNA
- Role
- root
- s
- Safety
- SCI
- Science
- Second
- security
- sensors
- September
- Sex
- Signal
- small
- some
- Spatial
- specific
- Stability
- stable
- State
- storage
- Strategy
- structural
- Study
- Switch
- syntax
- synthetic
- system
- Systems
- Target
- targeted
- techniques
- Technologies
- Technology
- The
- their
- three
- Through
- tightly
- time
- to
- tobacco
- Toolbox
- toolkit
- tools
- toward
- Transformation
- treatment
- Trends
- trial
- under
- use
- Vaccine
- vaccines
- Valuable
- via
- virus
- viruses
- volatile
- W
- Way..
- WELL
- Wellness
- West
- white
- with
- without
- X
- zephyrnet